NIfTI Image Annotation
For 3D Medical & Neuroscience AI
JTheta.ai supports full-featured annotation for NIfTI (.nii, .nii.gz) files — a must-have for teams
working with 3D volumetric data in radiology, neuroscience, and medical AI research. Our platform helps streamline the complex process of labeling MRI and fMRI datasets with accuracy, speed, and AI-assisted tools — all in a secure, browser-based environment.
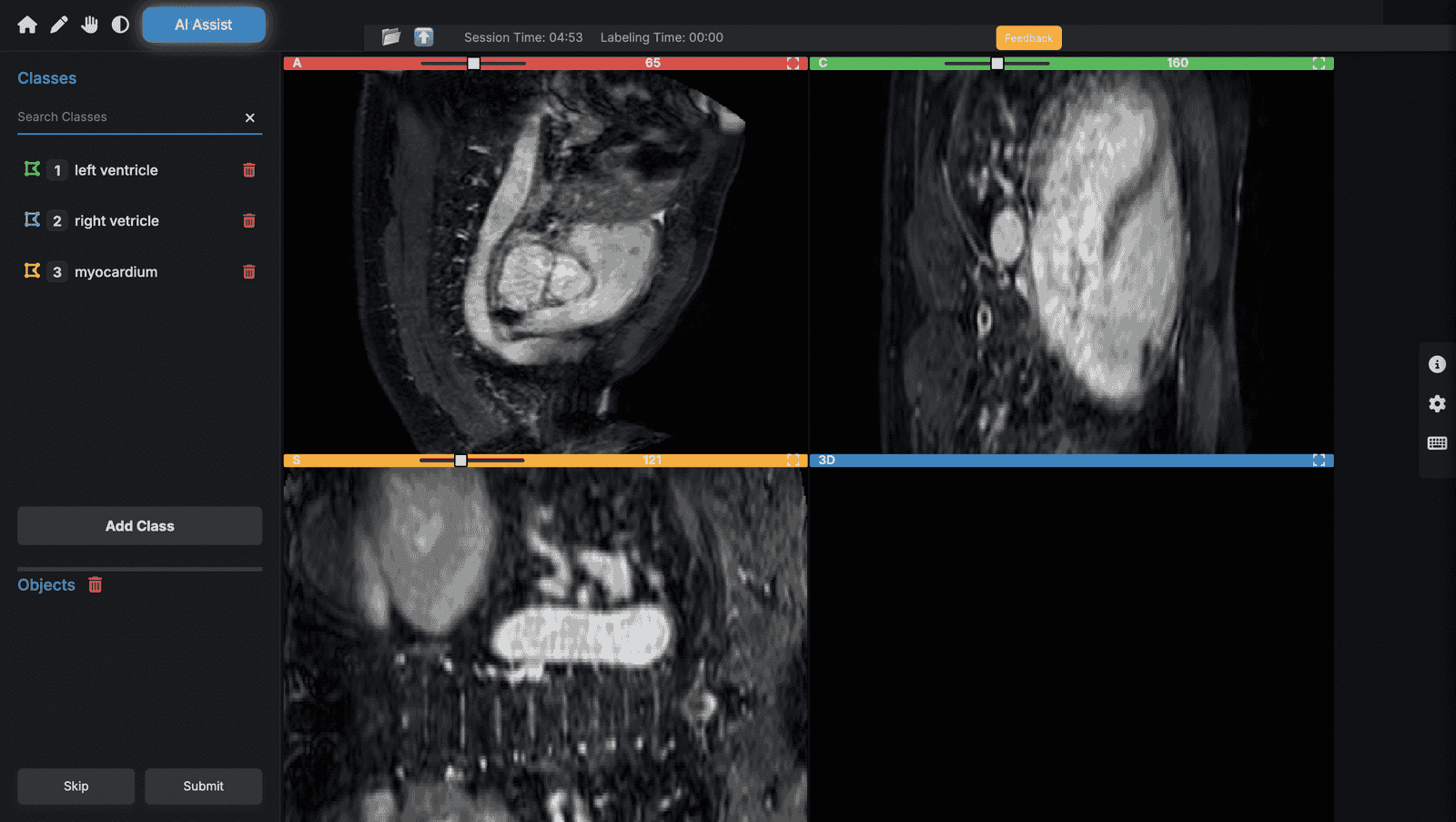
3D Volume Support for NIfTI Files
● Load .nii and .nii.gz files directly
● Navigate across slices in axial, sagittal, and coronal views
● Full volume rendering for precise region labeling
Voxel-Level Annotation
● Create 3D masks or segmentations at voxel-level precision
● Useful for tumor detection, brain region mapping, and structural analysis
Smart Tools for Complex Anatomy
● Polygon, brush, lasso tools for contouring
● Adjustable transparency and slice comparison
● Overlays for ground-truth and predicted masks
AI Pre-Annotation (Optional)
● Use AI to auto-label brain regions or anomalies
● Fine-tune manually with full control
Research-Ready Exports
● Compatible with AI training frameworks (e.g., MONAI, PyTorch)
● Export masks, coordinates, and metadata
Use Cases for NIfTI Annotation

Brain tumor segmentation for MRI
fMRI region labeling for neuroscience studies

3D cardiac structure labeling
Preoperative imaging analysis

Population-scale health studies using NIfTI datasets

Who Should Use It?
Why Use JTheta.ai for NIfTI Annotation?
- Faster 3D visualization
- Secure cloud access
- Built-in AI assistance
- Export-ready for ML training
FAQs
→ Yes, both .nii and .nii.gz formats are fully supported.
→ Absolutely. Our viewer supports plane switching and slice-based navigation.
→ No – JTheta.ai is 100% browser-based and secure.
